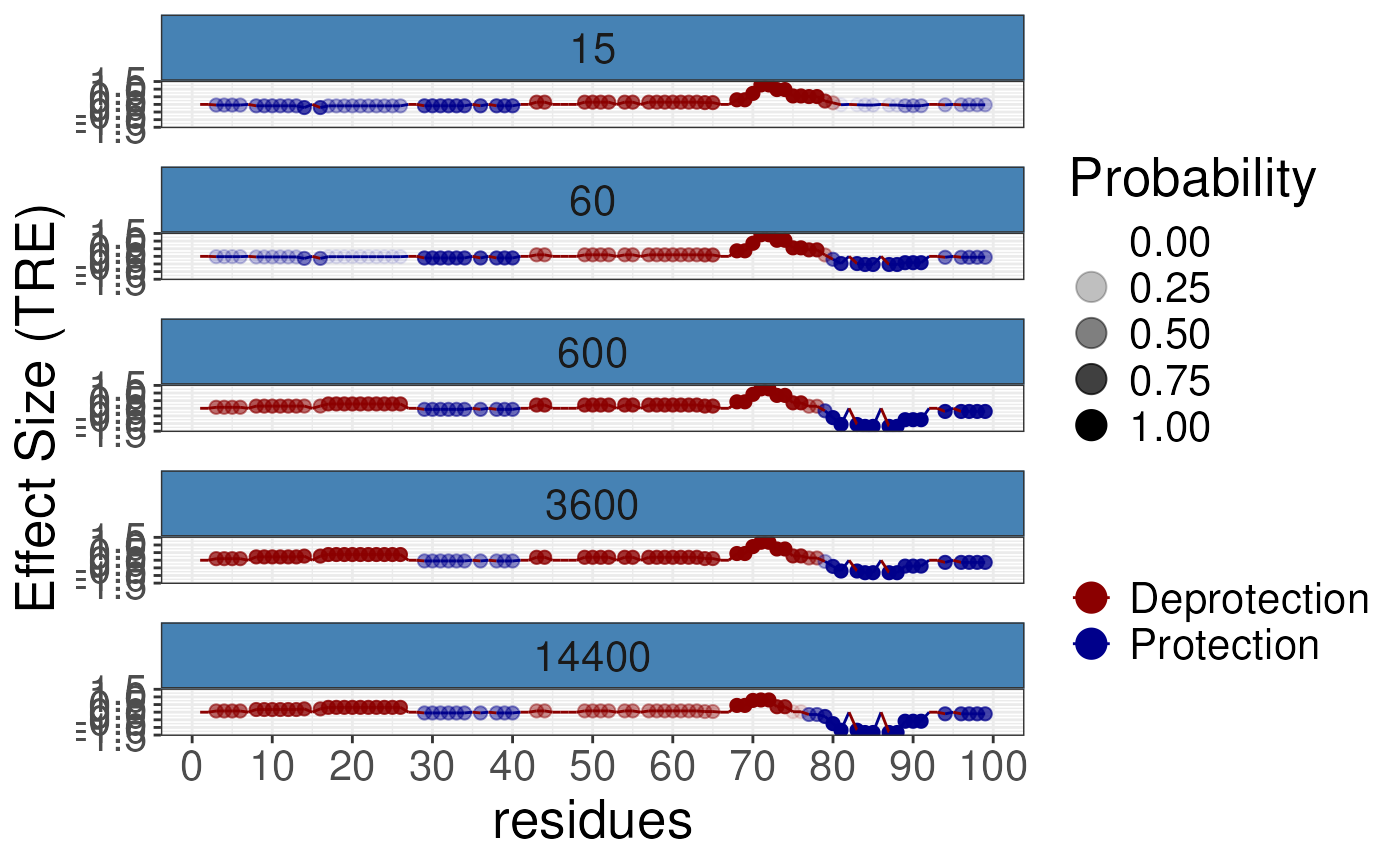
Rex butterfly plots for differential analysis
plotButterfly.RdRex butterfly plots for differential analysis
plotButterfly(diff_params, nrow = 5, quantity = "TRE", interval = NULL)Arguments
- diff_params
An object of class RexDifferential
- nrow
The number of rows in the facet (to seperate timepoints)
- quantity
The quantity to plot either "TRE" or "signedARE"
- interval
The interval to plot (Residues)
Value
Returns a ggplot object
Examples
require(RexMS)
require(dplyr)
require(ggplot2)
#> Loading required package: ggplot2
data("BRD4_apo")
data("BRD4_ibet")
BRD4_apo <- BRD4_apo %>% filter(End < 100)
BRD4_ibet <- BRD4_ibet %>% filter(End < 100)
numTimepoints <- length(unique(BRD4_apo$Exposure))
Timepoints <- unique(BRD4_apo$Exposure)
numPeptides <- length(unique(BRD4_apo$Sequence))
rex_test <- rex(HdxData = DataFrame(BRD4_ibet),
numIter = 10,
R = max(BRD4_ibet$End),
numtimepoints = numTimepoints,
timepoints = Timepoints,
seed = 1L,
tCoef = c(0, rep(1, 5)),
BPPARAM = SerialParam())
#> Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
#>
|
| | 0%
#> Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
#>
|
| | 0%
rex_test <- RexProcess(HdxData = DataFrame(BRD4_ibet),
params = rex_test,
range = 5:10,
thin = 1, whichChains = c(1,2))
rex_diff <- processDifferential(params = rex_test,
HdxData = DataFrame(BRD4_apo),
whichChain = c(1))
gg1 <- plotButterfly(diff_params = rex_diff,
nrow = 5,
quantity = "TRE")
print(gg1)