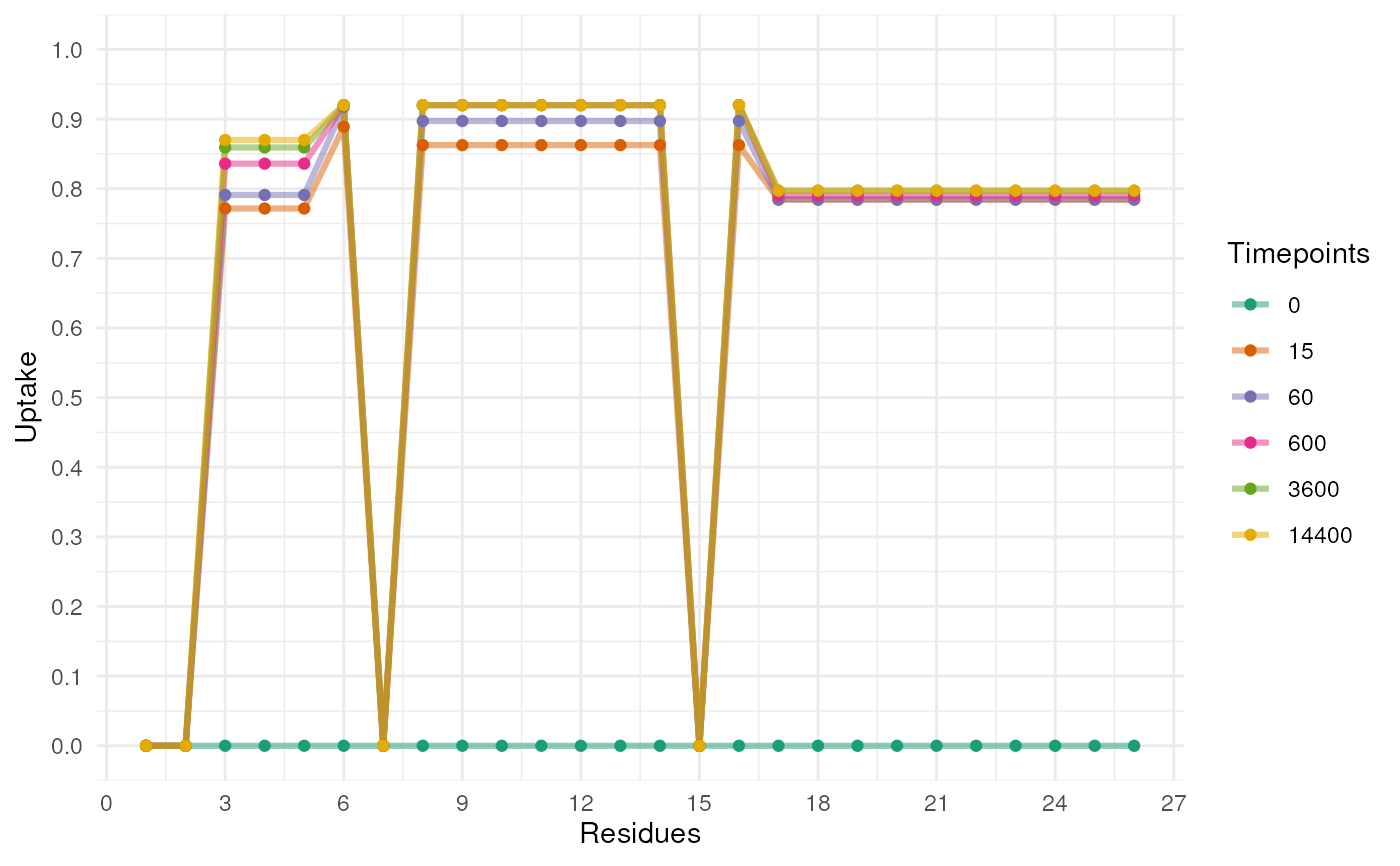
Plot the uptake from the output of a Rex model from the uptakePredict function
plotUptake.RdPlot the uptake from the output of a Rex model from the uptakePredict function
plotUptake(Uptake, facet = FALSE, values = brewer.pal(9, "Dark2"), nrow = 2)Arguments
- Uptake
An object of class DataFrame containing the results of the uptakePredict function
- facet
A logical value indicating whether to facet the plot (Facets by timepoints)
- values
A vector of colours to use in the plot of timepoints
- nrow
An integer value indicating the number of rows in the facet.
Value
Returns a ggplot object
Examples
require(RexMS)
require(dplyr)
data("BRD4_apo")
BRD4_apo <- BRD4_apo %>% filter(End < 40)
numTimepoints <- length(unique(BRD4_apo$Exposure))
Timepoints <- unique(BRD4_apo$Exposure)
rex_example <- rex(HdxData = DataFrame(BRD4_apo),
numIter = 4, # typically much larger
R = max(BRD4_apo$End),
numtimepoints = numTimepoints,
timepoints = Timepoints,
seed = 1L,
numChains = 1L,
tCoef = c(0, rep(1, 5)),
BPPARAM = SerialParam())
#> Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
#>
|
| | 0%
rex_example <- RexProcess(HdxData = DataFrame(BRD4_apo),
params = rex_example,
thin = 1,
range = 1:4,
whichChains = c(1))
Uptake <- uptakePredict(rex_example)
plotUptake(Uptake)
#> Warning: n too large, allowed maximum for palette Dark2 is 8
#> Returning the palette you asked for with that many colors